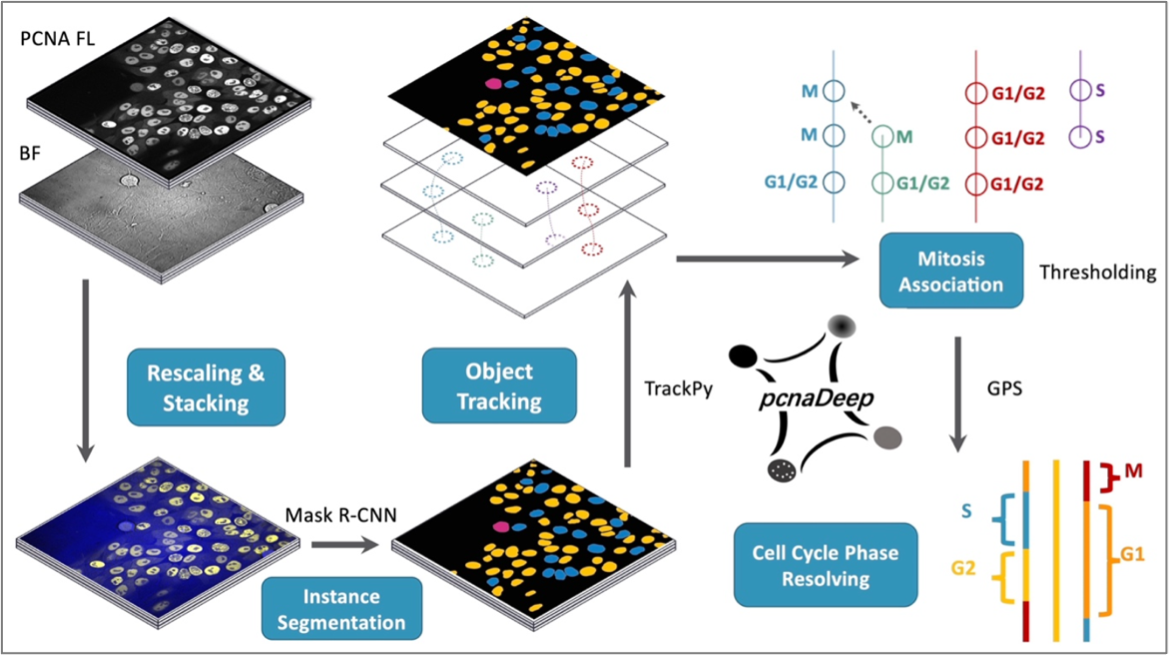
Quantitative single-cell analysis has revolutionised our approach to study the molecular events that lead to cell fate decisions. However, a common challenge when performing single-cell analysis is the limited computational tools available to track single-cells over multiple cell generations robustly. This is because when cells undergo cell division, they form daughter cells that can confound the algorithms used to track the cells, leading to high false mother-daughter assignments. This means extensive manual corrections are often required, making it a time-consuming practice.
In the recent publication in Bioinformatics, Kuan Yoow Chan's lab developed an application that uses deep learning to classify cells based on the pattern of the well-established fluorescently tagged PCNA protein. By using the PCNA pattern to identify the cell cycle state of cells, we were able to assign mother-daughter relationships rapidly and accurately. This novel approach has high accuracy rates and relatively low computational costs, allowing us to generate high-quality single-cell lineages from large long-term imaging datasets.
LINK:
https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btac602/6680181
About lab
Using genetic, cell biology and biochemical assays, Kuan Yoow Chan' Lab aim to study how the changes in centrosomal signalling lead to the deregulation of the cell cycle in cancer.







