Recently, Dr.Wanlu Liu from Zhejiang University-University of Edinburgh Institute and Prof. Steven E. Jacobsen from UCLA published an article, entitled Ectopic targeting of CG DNA methylation in Arabidopsis with the bacterial SssI methyltransferase, on Nature Communications. The study shows that targeting the CG-specific methyltransferase M.SssI with an artificial zinc finger protein to the promoter of the Arabidopsis FLOWERING WAGENINGEN (FWA) gene can establish heritable CG methylation and silencing of FWA in Arabidopsis (Figure 1). In addition, the author observes highly heritable widespread ectopic CG methylation mainly over euchromatic regions (Figure 2 A, B). By using the techniques including WGBS, ChIP-seq, ATAC-seq, etc., the author identified the Differentially methylated region (DMR), analyze the epigenetic marks on these DMRs, and study the relationship between the CG methylation and other epigenetic modifications. In addition, by analyzing the DMRs across multiple generations of Arabidopsis, the author studies the inheritance pattern of ectopic CG methylation.
Dr.Wanlu Liu is the first author and co-corresponding author of the article. The undergraduate student from Zhejiang University-University of Edinburgh Institute Yuxing Zhou participates in the research.
Abstract
DNA methylation is an evolutionarily conserved epigenetic modification that plays critical roles in regulating gene expression, silencing transposable elements, genomic imprinting, X-chromosome silencing, etc. In Arabidopsis, DNA methylation occurs within three sequence contexts: CG, CHG, and CHH (where H represents A, T, or C). The maintenance of CG methylation requires the DNA methyltransferase METHYLTRANSFERASE 1 (MET1, the homolog of mammalian DNMT1), while maintenance of CHG and CHH methylation redundantly requires CHROMOMETHYLASE 3 (CMT3), CMT2, and DRM2. Using techniques including zinc fingers (ZF), TAL, or CRISPR/Cas9 to target epigenetic marks like DNA methylation to specific loci is important in both basic research and in crop plant engineering. However, the heritability of targeted DNA methylation, how it impacts gene expression, and which epigenetic features are required for proper establishment is mostly unknown.
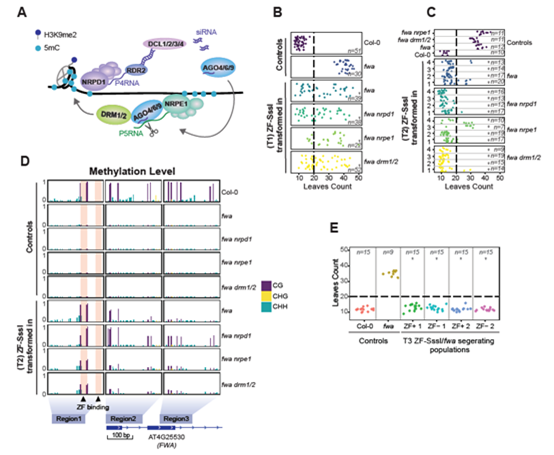
Figure 1
By comparing ectopic CG methylation and other epigenetic modification, the author finds that ectopic methylation occurs preferentially at less open chromatin that lacks positive histone marks. Although the ectopic CG methylation is enriched in the gene body region, this hypermethylation shows little effect on transcription (Figure 2 C-E). The author also observes that the ectopic methylation triggers a mild but significant reduction in the accumulation of H2A.Z and H3K27me3. By analyzing through four generations of Arabidopsis, the author observes the significant heritability of the CG methylation region. These results outline general principles of the heritability and interaction of CG methylation with other epigenomic features that should help guide future efforts to engineer epigenomes.

Figure 2







