Organelles engage in signal transduction and material exchange through membrane contact sites, which play significant roles in lipid synthesis, calcium ion transfer, and autophagy. Mitochondria have been widely reported to form contact sites with the endoplasmic reticulum (ER), referred to as mitochondria-ER contact sites (MERCs). Early studies suggested that mitochondrial fusion proteins Mitofusin 1 and 2 (MFN1/2) function as physical linkers at these contact sites by forming dimers with MFN2 on the ER. However, super-resolution imaging of MFN2 fails to detect MFN2 signal on ER membranes. Thus, the model that ER-MFN2 dimerizes with mitochondrial-MFN2 requires re-examination.
Recent findings propose that MFN2 specifically transports phosphatidylserine (PS) from the ER to mitochondria for the synthesis of phosphatidylethanolamine (PE), further indicating MFN2's potential involvement in multiple functions at MERCs. Whether all these functions are mediated by MFN2-MFN2 dimers and whether other unknown ER proteins participate with MFN2 remains unresolved. Therefore, identifying new ER protein partner for MFN2 and elucidating their biological functions will help in reevaluating the role of MFN2-MFN2 homodimers in mediating MERCs and functions related.
On 12 August 2024, Dr. Zhi Hong's group at ZJE published an article titled "Dynamic interaction of REEP5-MFN1/2 enables mitochondrial hitchhiking on tubular ER" in the Journal of Cell Biology. This study is the first to reveal that the ER-shaping protein REEP5, as a novel interacting partner of MFN1/2, mediates the mitochondrial hitchhiking on the tubular ER, ultimately achieving uniform mitochondrial distribution throughout cytoplasm and regulating cellular reactive oxygen species (ROS) homeostasis.

Previous results indicated that knockdown or knockout of REEP family members in the ER leads to mitochondrial dysfunction. Thus the study initially performed a small-scale screening of the REEP protein family, discovering that REEP5 specifically binds MFN1/2. Live-cell imaging further confirmed the enrichment of REEP5 and MFN1/2 at MERCs. The Using GI-SIM super-resolution live imaging, ER and mitochondria displayed a dynamic “association-dissociation” pattern, with REEP5-MFN2 tethers driving mitochondrial hitchhiking on the tubular ER. The study found that weakening REEP5-MFNs interactions leads to mitochondrial collapse to the perinuclear region. Interestingly, when REEP5-MFNs interactions are locked, mitochondrial can no longer be randomly released from tubular ER, they remained attached to the ER instead and resulting in mitochondrial hyperfusion. The findings suggest that the dynamic binding between REEP5 and MFN1/2 underlies the molecular mechanism of dynamic ER-mitochondria contact.
Previous studies have shown that the subcellular localization of mitochondria is closely related to mitochondrial ROS homeostasis. Using the mitochondrial-localized ROS sensors roGFP2 and HyPer7, the researchers found that mitochondrial ROS level is tightly regulated by REEP5-MFN1/2 tethering strengths, e.g. mitochondrial ROS is upregulated once REEP5-MFN1/2 tethering is disrupted.
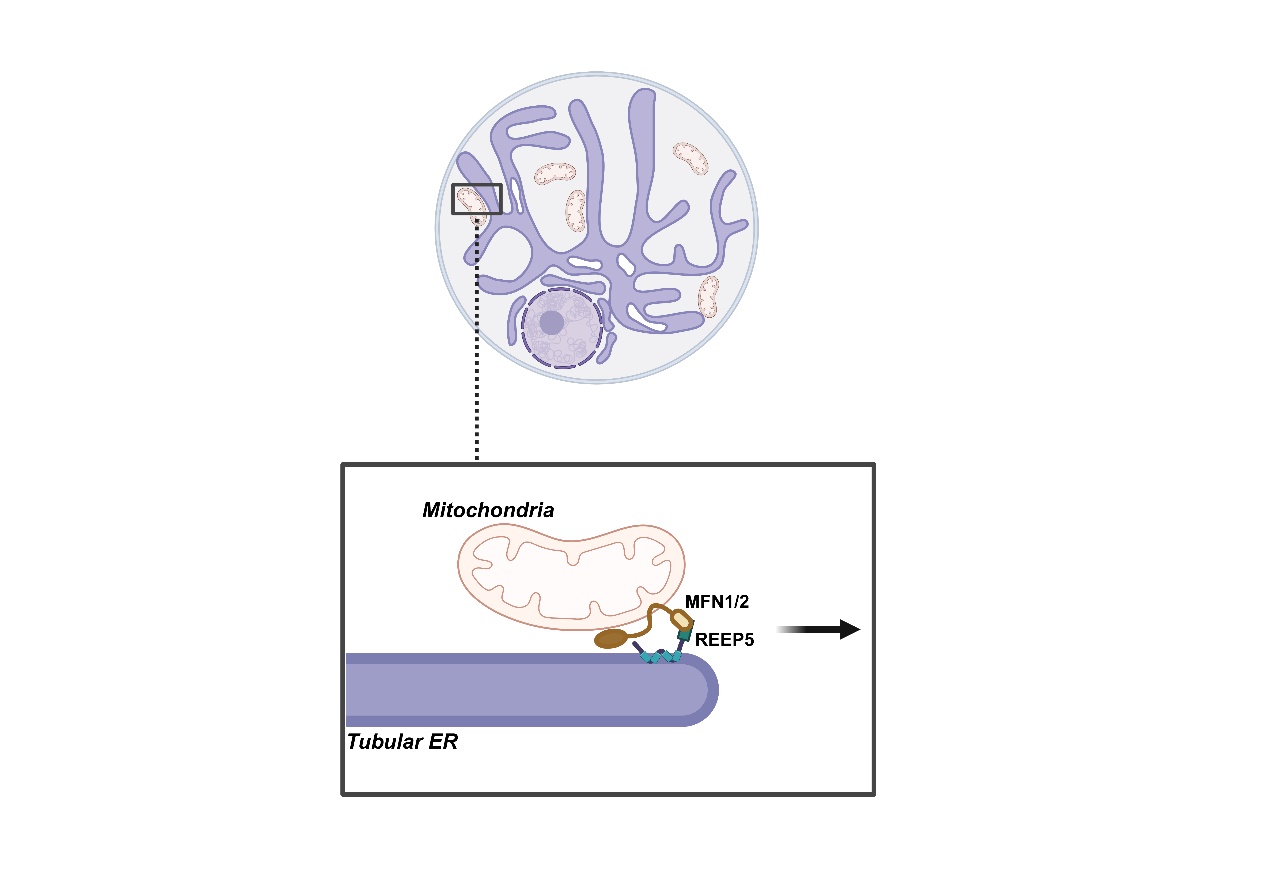
In summary, this study elucidates a novel mechanism by which the REEP5-MFN1/2 protein complex facilitates mitochondrial "hitchhiking" along the tubular ER, thereby promoting uniform mitochondrial distribution within the cell and regulating cellular ROS homeostasis. The findings offer theoretical evidence suggesting that organelle interactions are intrinsically dynamic processes featured by cycles of association and dissociation.
Dr. Zhi Hong from ZJE serves as the corresponding author, with Dr. Shue Chen and Ph.D. candidate Yang Sun as co-first authors. Additionally, Ph.D. candidates Lan Yang and Yuling Qin from ZJE contributed significantly to this study. This research received support from the National Natural Science Foundation of China.
Link to the paper:
https://doi.org/10.1083/jcb.202304031
About Dr. Zhi Hong’ lab
Dr. Zhi Hong’s research group focuses on organelle interactions and cellular homeostasis, as well as investigating the molecular mechanisms and biological functions related to lipid metabolism. Their findings have been published in esteemed journals such as the Journal of Cell Biology, Cell Research, Autophagy, and Antiviral Research.







