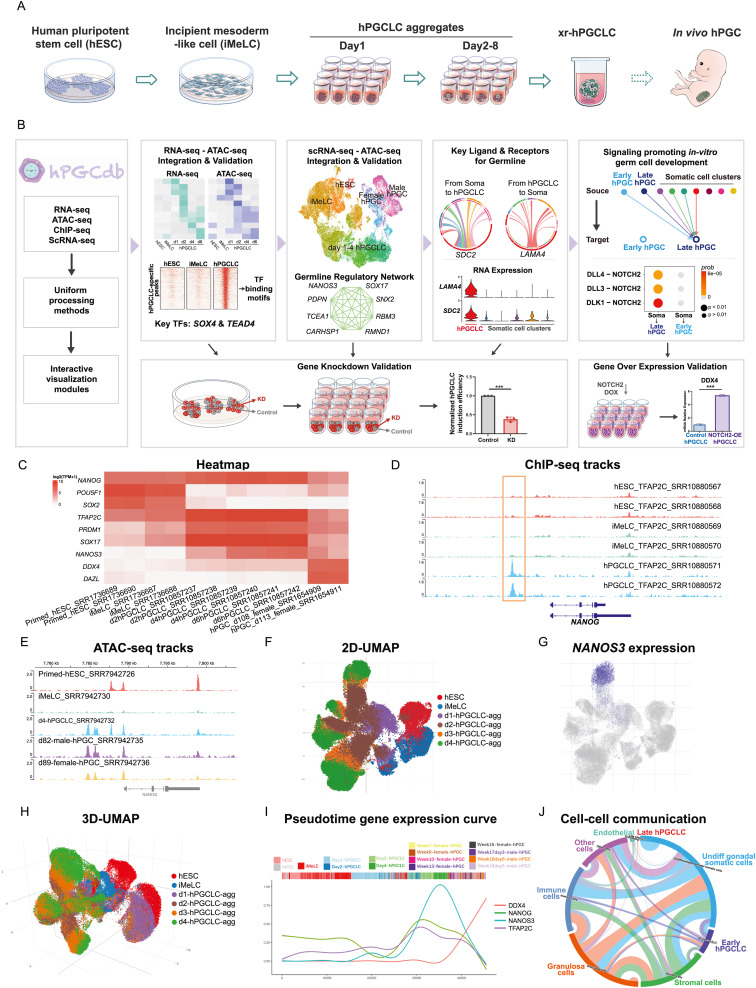
This paper details an innovative study on the development of human primordial germ cells (PGCs), the precursors to the germline, specified early in embryonic life. Despite the significance of PGCs in reproductive biology, their developmental pathways have remained largely unexplored due to the ethical and technical challenges associated with human embryonic research.
To address these gaps, we established a comprehensive human PGC database (hPGCdb), which consists of 581 RNA-seq, 54 ATAC-seq, 45 ChIP-seq, and 69 single-cell RNA-seq samples from various stages of PGC development. This database serves as a pivotal resource for understanding the regulatory mechanisms governing human PGC specification and differentiation.
Through integrated multi-omics analysis, we identified several key transcription factors and regulatory networks that control germ cell fate, highlighting the differences between human and model organisms like mice. We further validated the functional importance of specific transcription factors using Gene KnockDown and KnockOut, confirming their roles in germ cell development.
Additionally, our research explored the soma-germline interaction network, revealing roles for SDC2 and LAMA4 in PGC development and the influence of soma-derived NOTCH2 signaling in germ cell differentiation. These findings enhance our understanding of the extrinsic factors influencing germ cell fate and open new avenues for research into reproductive health and disease.
In summary, this study not only provides a detailed map of the regulatory landscape governing human PGC development but also introduces a valuable genomic resource that enhances our understanding of human reproductive biology and developmental genetics.