A. Professor Irving's laboratory at ZJU-UoE Institute, in collaboration with the Sloan Lab, University of Edinburgh, and the Compton laboratory, National Cancer Institute USA, have published an article on the chinese horseshoe bat IFITM3 antiviral protein in PLOS Pathogens. https://doi.org/10.1371/journal.ppat.1012763
Zoonotic transmission occurs when viruses ‘jump’ from animals into human,potentially leading to viral outbreaks such as the COVID-19 pandemic, which poses a significant threat to public health. Bats serve as the origin of many zoonotic viruses; their unique immune system may allow them to harbor these pathogens without developing diseases.The human interferon-induced transmembrane protein (IFITM) family are critical antiviral components that have been shown to influence the pathogenesis of viral infections. They restrict virus entry and are key players in antiviral interferon responses. In addition, the unique IFITM repertoires in different species influence their resistance to viral infections, but the role of IFITMs in shaping the enhanced antiviral immunity of reservoir bat species is unclear. Thus characterization of bat IFITMs is needed to identify factors that predispose species to act as viral reservoirs.
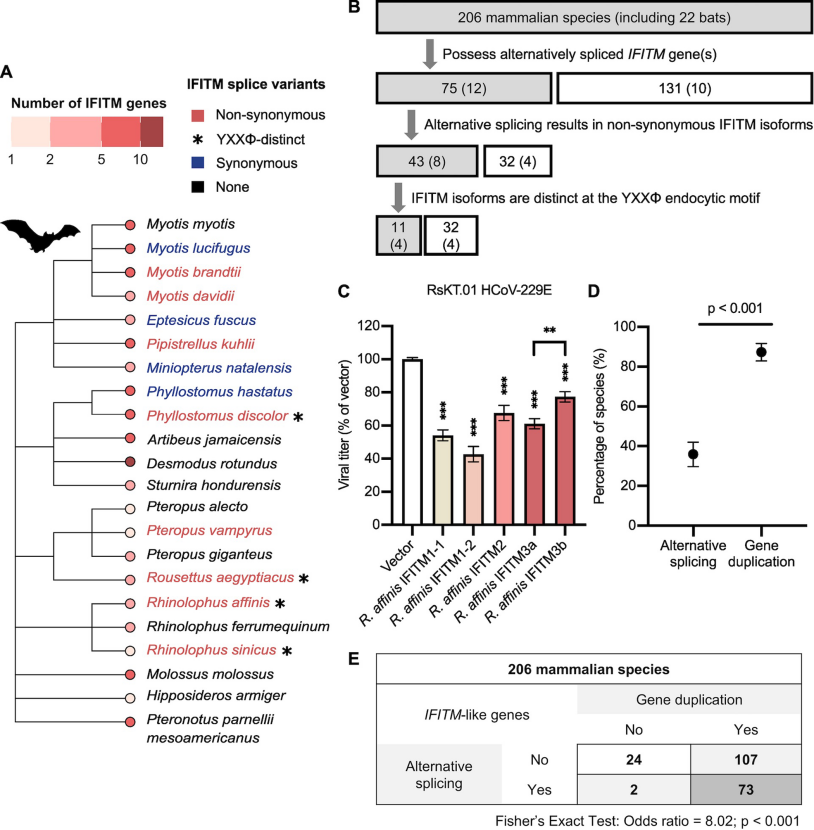
Here, we identified an IFITM gene in Chinese rufous horseshoe bat, a natural host of severe acute respiratory syndrome (SARS)-related coronaviruses, that is alternatively spliced to produce two IFITM isoforms in native cells as shown by transcriptomics. These bat IFITMs have conserved structures in vitro as demonstrated by circular dichroism spectroscopy, yet they exhibit distinct antiviral specificities against influenza A virus, Nipah virus and coronaviruses including SARS-CoV, SARS-CoV-2 and MERS-CoV. In parallel with human IFITM1-3, bat IFITM isoforms localize to distinct sites of virus entry which influences their antiviral potency.
Further bioinformatic analysis of IFITM repertoires in 206 mammals reveals that alternative splicing is a recurring strategy for IFITM diversification, albeit less widely adopted than gene duplication. 13% of sampled mammals have only one predicted IFITM transcript. These findings demonstrate that alternative splicing is a key strategy for evolutionary diversification in the IFITM family. Altogether, our studyhighlights an example of convergent evolution where species-specific selection pressures led to expansion of the IFITM family through multiple means, underscoring the importance of IFITM diversity as a component of innate immunity.







