Cancer, a disease that strikes fear into the hearts of countless people, is essentially a "genetic war" that occurs within the human body. The gradual accumulation of genetic mutations within cells leads to a loss of normal growth and division control, thereby influencing the onset, progression, maintenance, spread, and metastasis of cancer.
Studying genomic mutations not only helps us better understand the biological mechanisms of cancer but also provides crucial clues for developing new treatment methods. Scientists have been researching these mutations in the hope of finding breakthroughs to combat cancer.
This study discovered a direct link between synonymous mutations and the epitranscriptome, revealing a novel regulatory mechanism in tumorigenesis and progression, offering new insights for precision medicine and targeted drug therapy in oncology. Additionally, this discovery expands our understanding of the "central dogma" of molecular biology—genomic DNA sequences can achieve fine regulation by directly influencing mRNA modifications.
The concept of gene mutations is familiar to many, with those that promote cancer development being termed "driver mutations." If genes are likened to a complex "book of life," then gene mutations are akin to "errors" in the book: some are "typos," others are "misordered paragraphs," and perhaps some are "omissions." Sometimes, these errors do not affect "reading," but other times, they can drastically alter the story of the "book of life."
Among these, point mutations (changes in a single base pair) are a very common and crucial type of driver mutation in cancer. Existing research shows that a single tumor typically acquires 2 to 8 driver mutations, most of which are point mutations.
Among these point mutations, there is a type called "synonymous mutations," which account for a significant proportion, about 25-30%. These do not alter the amino acid sequence of proteins and seem to have no impact on protein function, appearing unrelated to tumorigenesis.
At this point, a chemical modification occurring on RNA molecules caught the team's attention — epitranscriptomic modifications. These modifications do not change the nucleotide sequence of RNA but can affect RNA's structure, stability, and function. Recent extensive research has shown that epitranscriptomic modifications, especially RNA N6-methyladenosine (m6A) modifications, play a key role in the onset and progression of cancer. For example, abnormal m6A modifications have been found in acute myeloid leukemia and glioblastoma.
Based on these clues, the research team hypothesized that the seemingly "harmless" synonymous mutations accumulated in cancer cells might be linked to "epitranscriptomic modifications".
In October 2022, the laboratory officially launched the project.
First, they systematically quality-controlled and organized the human pan-cancer genomic mutations using the latest COSMIC database and TCGA data, while also collecting and organizing the latest human m6A data (m6A-Atlas v2.0 and REPIC database, etc.). Through three stringent screening criteria (Figure 1, including mutation reproducibility screening, m6A data coverage, and further reliability screening), they ultimately identified 12,849 potential m6A Disruption Mutations (m6A-DMs). These mutations can be further divided into synonymous mutations (sm6A-DM) and missense mutations (mm6A-DM).
They found that synonymous mutations (sm6A-DM) are more likely to occur in tumor suppressor genes. Tumor suppressor genes inhibit abnormal cell proliferation; once these genes mutate or lose function, cells may lose control over proliferation, leading to tumor formation.
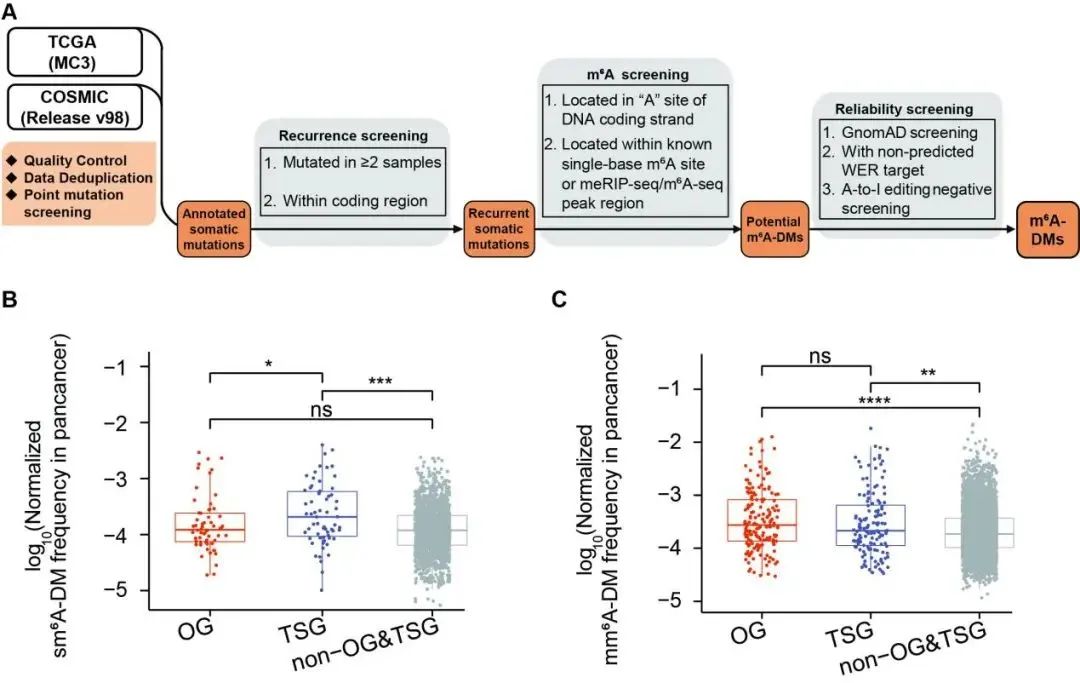
Figure 1. The identification process of m6A-DM and the distribution characteristics of genes.
Subsequently, based on the analysis of cancer genomic mutation frequencies, the researchers identified two top-ranked sm6A-DM sites (CDKN2A-c.A294B and BRCA2-c.A1365G) among the more than ten thousand potential disruptive mutations and conducted functional validation and mechanistic exploration.
They employed previously developed regulatory tools (dCasRx site-specific methylation editing system) combined with knock-in based site-specific mutation technology to regulate highly associated tumor type cells at both the epitranscriptomic modification level and the genomic in situ level. Experimental results showed that the m6A modification levels at these two sites were positively correlated with the abundance of corresponding gene transcripts, meaning that higher m6A modification levels at these sites corresponded to higher mRNA quantities, and vice versa. When mutations occurred at these sites, they disrupted the m6A modifications and affected mRNA stability, leading to a decrease in the corresponding gene's mRNA quantity and affecting gene expression levels (Figure 2).
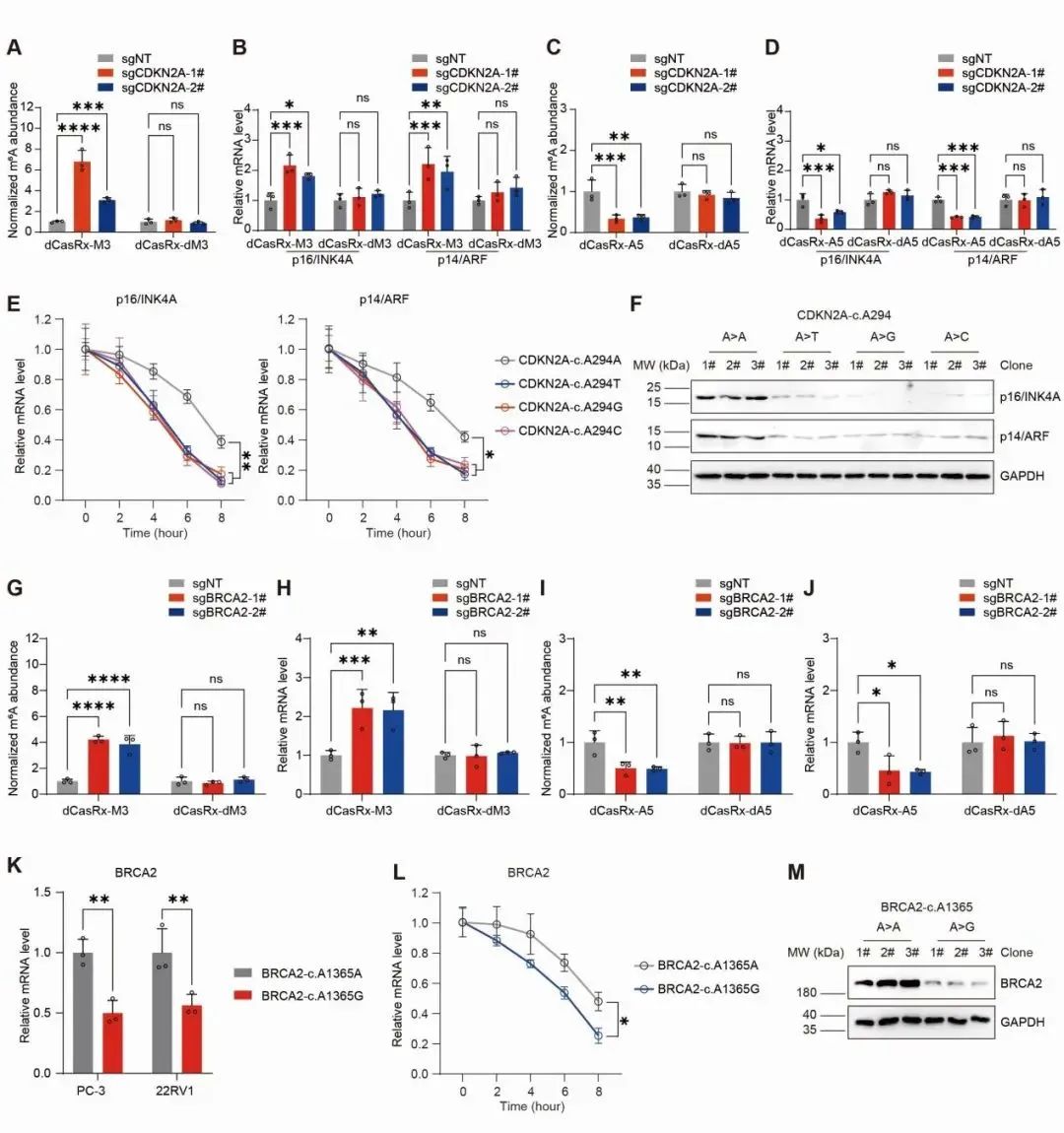
Figure 2.The significant impact of sm6A-DM on mRNA stability.
Returning to our initial topic, how do these findings relate to the onset and progression of cancer? As mentioned earlier, epitranscriptomic modifications play a key role in tumorigenesis and progression. Since synonymous mutations can affect epitranscriptomic modifications, they might similarly influence tumorigenesis and progression.
Through experimental analysis, the research team found that a mutation at the CDKN2A-c.A294B site led to a significant downregulation of CDKN2A transcript and protein levels, accelerating the tumor cell division cycle, thereby enhancing tumor cell proliferation and tumor formation speed in mice, greatly increasing the malignancy of the tumor.
At this point, a novel regulatory mechanism for tumorigenesis and progression became clear—sm6A-DMs genomic synonymous mutations can disrupt normal m6A modifications of mRNA, weaken mRNA stability, cause changes in mRNA and protein abundance levels, and alter the tumorigenesis and progression process.
It turns out that synonymous mutations are not entirely "harmless"!
Based on the rigor of scientific research, the team further advanced these results—clinically, an inhibitor called Olaparib has been approved by the FDA for treating tumors with BRCA1 and BRCA2 defects. The research team designed a set of experiments to validate the implications of their findings for tumor diagnosis and treatment. As expected, tumor cells with the BRCA2-c.A1365G mutation were more sensitive to PARPi drug Olaparib treatment, and tumor growth was well suppressed in tumor-bearing mice with this sm6A-DM mutation after Olaparib treatment. This discovery provides new insights for precision medicine and targeted drug therapy in oncology. In the future, more detection and targeted therapies for sm6A-DMs may emerge, allowing for more precise identification and treatment of specific types of cancer.
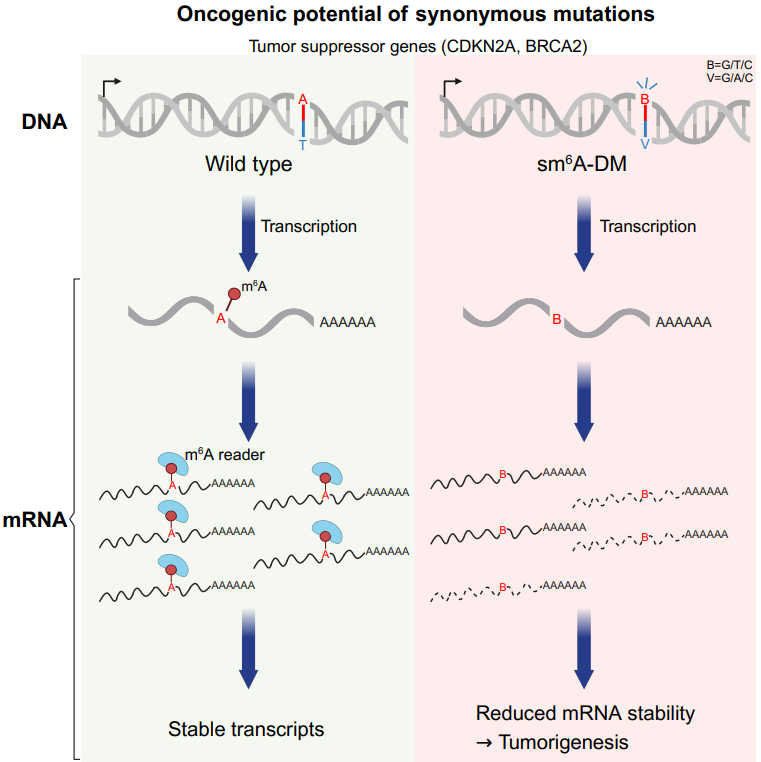
Figure 3. The role of sm6A-DM in the process of tumorigenesis and development.
Moreover, this study holds a more significant meaning, revealing a previously overlooked dimension of the central dogma of molecular biology: DNA mutations can directly determine mRNA modification patterns. The familiar "central dogma" posits that DNA information is transcribed into mRNA, which is then translated into proteins. This study further expands this dogma, indicating that DNA mutations can directly influence mRNA modifications, thereby finely regulating gene expression, enriching our understanding of the essence of life.
Dr. Di Chen from the Zhejiang University-University of Edinburgh Institute, Dr. Qi Xie and Dr. Yanmei Dou from Westlake Laboratory are the co-corresponding authors of this study. Qizhe Shao, a Ph.D. student at the Zhejiang University-University of Edinburgh Institute, and Yiheng Lan, an assistant researcher in Qi Xie's team, and Zhen Xia, a Ph.D. student, are the co-first authors of this paper. This research received funding support from the National Natural Science Foundation of China, the Major Projects of Technological Innovation 2030, the Zhejiang "Pioneer" and "Leading Goose" R&D Program, and the Westlake Education Foundation.







